Difference between revisions of "Protein"
| Line 1: | Line 1: | ||
| − | <p><strong>Proteins</strong> are large | + | <p><strong>Proteins</strong> are large organic compounds made of amino acids arranged in a linear chain and joined together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. The sequence of amino acids in a protein is defined by a gene and encoded in the genetic code. Although this genetic code specifies 20 "standard" amino acids, the residues in a protein are often chemically altered in post-translational modification: either before the protein can function in the cell, or as part of control mechanisms. Proteins can also work together to achieve a particular function, and they often associate to form stable complexes.</p> |
| − | <p>Like other biological | + | <p>Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of all living organisms and participate in every process within cells. Many proteins are enzymes that catalyze biochemical reactions, and are vital to metabolism. Other proteins have structural or mechanical functions, such as the proteins in the cytoskeleton, which forms a system of scaffolding that maintains cell shape. Proteins are also important in cell signaling, immune responses, <font color="#810081">cell adhesion</font>, and the cell cycle. Protein is also a necessary component in our diet, since animals cannot synthesise all the amino acids and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that can be used for protein synthesis.</p> |
| − | <p>The word <em>protein</em> comes from the | + | <p>The word <em>protein</em> comes from the Greek <em>πρώτα</em> ("prota"), meaning "<em>of primary importance</em>" and these molecules were first described and named by Jöns Jakob Berzelius in 1838. However, proteins' central role in living organisms was not fully appreciated until 1926, when James B. Sumner showed that the enzyme urease was a protein.<sup class="reference" id="_ref-0">[1]</sup> The first protein to be sequenced was insulin, by Frederick Sanger, who won the Nobel Prize for this achievement in 1958. The first protein structures to be solved included haemoglobin and myoglobin, by Max Perutz and Sir John Cowdery Kendrew, respectively, in 1958.<sup class="reference" id="_ref-1">[2]</sup><sup class="reference" id="_ref-2">[3]</sup> Both proteins' three-dimensional structures were first determined by x-ray diffraction analysis; the structures of myoglobin and haemoglobin won the 1962 Nobel Prize in Chemistry for their discoverers.</p> |
| − | |||
<p><a id="Biochemistry" name="Biochemistry"></a></p> | <p><a id="Biochemistry" name="Biochemistry"></a></p> | ||
<h2><span class="mw-headline">Biochemistry</span></h2> | <h2><span class="mw-headline">Biochemistry</span></h2> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main articles: | + | <div class="noprint"><em>Main articles: Amino acid and peptide bond</em></div> |
</dd></dl> | </dd></dl> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 302px | + | <div class="thumbinner" style="WIDTH: 302px"><img class="thumbimage" height="65" alt="Resonance structures of the peptide bond that links individual amino acids to form a protein polymer." width="300" longdesc="/wiki/Image:Peptide_group_resonance.png" src="http://upload.wikimedia.org/wikipedia/en/thumb/1/17/Peptide_group_resonance.png/300px-Peptide_group_resonance.png" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | + | Resonance structures of the peptide bond that links individual amino acids to form a protein polymer.</div> | |
</div> | </div> | ||
</div> | </div> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 302px | + | <div class="thumbinner" style="WIDTH: 302px"><img class="thumbimage" height="211" alt="Section of a protein structure showing serine and alanine residues linked together by peptide bonds. Carbons are shown in white and hydrogens are omitted for clarity." width="300" longdesc="/wiki/Image:Peptide_bond.jpg" src="http://upload.wikimedia.org/wikipedia/en/thumb/b/ba/Peptide_bond.jpg/300px-Peptide_bond.jpg" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
Section of a protein structure showing serine and alanine residues linked together by peptide bonds. Carbons are shown in white and hydrogens are omitted for clarity.</div> | Section of a protein structure showing serine and alanine residues linked together by peptide bonds. Carbons are shown in white and hydrogens are omitted for clarity.</div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <p>Proteins are linear polymers built from 20 different L-α- | + | <p>Proteins are linear polymers built from 20 different L-α-amino acids. All amino acids share common structural features including an α carbon to which an amino group, a carboxyl group, and a variable side chain are bonded. Only proline differs from this basic structure, as it contains an unusual ring to the N-end amine group, which forces the CO-NH amide moiety into a fixed conformation.<sup class="reference" id="_ref-3">[4]</sup> The side chains of the standard amino acids, detailed in the list of standard amino acids, have different chemical properties that produce proteins' three-dimensional structure and are therefore critical to protein function. The amino acids in a polypeptide chain are linked by peptide bonds formed in a dehydration reaction. Once linked in the protein chain, an individual amino acid is called a <em>residue</em> and the linked series of carbon, nitrogen, and oxygen atoms are known as the <em>main chain</em> or <em>protein backbone</em>. The peptide bond has two resonance forms that contribute some double bond character and inhibit rotation around its axis, so that the alpha carbons are roughly coplanar. The other two dihedral angles in the peptide bond determine the local shape assumed by the protein backbone.</p> |
| − | <p>Due to the chemical structure of the individual amino acids, the protein chain has directionality. The end of the protein with a free carboxyl group is known as the | + | <p>Due to the chemical structure of the individual amino acids, the protein chain has directionality. The end of the protein with a free carboxyl group is known as the C-terminus or carboxy terminus, while the end with a free amino group is known as the N-terminus or amino terminus.</p> |
| − | <p>There is some ambiguity between the usage of the words <em>protein</em>, <em | + | <p>There is some ambiguity between the usage of the words <em>protein</em>, <em>polypeptide</em>, and <em>peptide</em>. <em>Protein</em> is generally used to refer to the complete biological molecule in a stable conformation, while <em>peptide</em> is generally reserved for a short amino acid oligomers often lacking a stable 3-dimensional structure. However, the boundary between the two is ill-defined and usually lies near 20-30 residues.<sup class="reference" id="_ref-Lodish_0">[5]</sup> <em>Polypeptide</em> can refer to any single linear chain of amino acids, usually regardless of length, but often implies an absence of a single defined conformation.</p> |
<p><a id="Synthesis" name="Synthesis"></a></p> | <p><a id="Synthesis" name="Synthesis"></a></p> | ||
<h2><span class="mw-headline">Synthesis</span></h2> | <h2><span class="mw-headline">Synthesis</span></h2> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main article: | + | <div class="noprint"><em>Main article: Protein biosynthesis</em></div> |
</dd></dl> | </dd></dl> | ||
| − | <div class="floatleft"><span | + | <div class="floatleft"><span><img height="145" alt="" width="125" longdesc="/wiki/Image:One_gene%2C_one_enzyme.gif" src="http://upload.wikimedia.org/wikipedia/en/4/4e/One_gene%2C_one_enzyme.gif" /></span></div> |
| − | <p>Proteins are assembled from amino acids using information encoded in | + | <p>Proteins are assembled from amino acids using information encoded in genes. Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called codons and each three-nucleotide combination stands for an amino acid, for example AUG stands for methionine. Because DNA contains four nucleotides, the total number of possible codons is 64; hence, there is some redundancy in the genetic code and some amino acids are specified by more than one codon. Genes encoded in DNA are first transcribed into pre-messenger RNA (mRNA) by proteins such as RNA polymerase. Most organisms then process the pre-mRNA (also known as a <em>primary transcript</em>) using various forms of post-transcriptional modification to form the mature mRNA, which is then used as a template for protein synthesis by the ribosome. In prokaryotes the mRNA may either be used as soon as it is produced, or be bound by a ribosome after having moved away from the nucleoid. In contrast, eukaryotes make mRNA in the cell nucleus and then translocate it across the nuclear membrane into the cytoplasm, where protein synthesis then takes place. The rate of protein synthesis is higher in prokaryotes than eukaryotes and can reach up to 20 amino acids per second.<sup class="reference" id="_ref-Dobson_0">[6]</sup></p> |
| − | <p>The process of synthesizing a protein from an mRNA template is known as | + | <p>The process of synthesizing a protein from an mRNA template is known as translation. The mRNA is loaded onto the ribosome and is read three nucleotides at a time by matching each codon to its base pairing anticodon located on a transfer RNA molecule, which carries the amino acid corresponding to the codon it recognizes. The enzyme aminoacyl tRNA synthetase "charges" the tRNA molecules with the correct amino acids. The growing polypeptide is often termed the <em>nascent chain</em>. Proteins are always biosynthesized from N-terminus to C-terminus.</p> |
| − | <p>The size of a synthesized protein can be measured by the number of amino acids it contains and by its total | + | <p>The size of a synthesized protein can be measured by the number of amino acids it contains and by its total molecular mass, which is normally reported in units of <em>daltons</em> (synonymous with atomic mass units), or the derivative unit kilodalton (kDa). Yeast proteins are on average 466 amino acids long and 53 kDa in mass.<sup class="reference" id="_ref-Lodish_1">[5]</sup> The largest known proteins are the titins, a component of the muscle sarcomere, with a molecular mass of almost 3,000 kDa and a total length of almost 27,000 amino acids.<sup class="reference" id="_ref-4">[7]</sup></p> |
<p><a id="Chemical_synthesis" name="Chemical_synthesis"></a></p> | <p><a id="Chemical_synthesis" name="Chemical_synthesis"></a></p> | ||
<h3><span class="mw-headline">Chemical synthesis</span></h3> | <h3><span class="mw-headline">Chemical synthesis</span></h3> | ||
| − | <p>Short proteins can also be synthesized chemically by a family of methods known as | + | <p>Short proteins can also be synthesized chemically by a family of methods known as peptide synthesis, which rely on organic synthesis techniques such as chemical ligation to produce peptides in high yield.<sup class="reference" id="_ref-5">[8]</sup> Chemical synthesis allows for the introduction of non-natural amino acids into polypeptide chains, such as attachment of fluorescent probes to amino acid side chains.<sup class="reference" id="_ref-6">[9]</sup> These methods are useful in laboratory biochemistry and cell biology, though generally not for commercial applications. Chemical synthesis is inefficient for polypeptides longer than about 300 amino acids, and the synthesized proteins may not readily assume their native tertiary structure. Most chemical synthesis methods proceed from C-terminus to N-terminus, opposite the biological reaction.</p> |
<p><a id="Structure_of_proteins" name="Structure_of_proteins"></a></p> | <p><a id="Structure_of_proteins" name="Structure_of_proteins"></a></p> | ||
<h2><span class="mw-headline">Structure of proteins</span></h2> | <h2><span class="mw-headline">Structure of proteins</span></h2> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main article: | + | <div class="noprint"><em>Main article: Protein structure</em></div> |
</dd></dl> | </dd></dl> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 502px | + | <div class="thumbinner" style="WIDTH: 502px"><img class="thumbimage" height="200" alt="Three possible representations of the three-dimensional structure of the protein triose phosphate isomerase. Left: all-atom representation colored by atom type. Middle: simplified representation illustrating the backbone conformation, colored by secondary structure. Right: Solvent-accessible surface representation colored by residue type (acidic residues red, basic residues blue, polar residues green, nonpolar residues white)." width="500" longdesc="/wiki/Image:Proteinviews-1tim.png" src="http://upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Proteinviews-1tim.png/500px-Proteinviews-1tim.png" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | Three possible representations of the three-dimensional structure of the protein | + | Three possible representations of the three-dimensional structure of the protein triose phosphate isomerase. Left: all-atom representation colored by atom type. Middle: simplified representation illustrating the backbone conformation, colored by secondary structure. Right: Solvent-accessible surface representation colored by residue type (acidic residues red, basic residues blue, polar residues green, nonpolar residues white).</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>Most proteins | + | <p>Most proteins fold into unique 3-dimensional structures. The shape into which a protein naturally folds is known as its native state. Although many proteins can fold unassisted simply through the structural propensities of their component amino acids, others require the aid of molecular chaperones to efficiently fold to their native states. Biochemists often refer to four distinct aspects of a protein's structure:</p> |
<ul> | <ul> | ||
| − | <li><em | + | <li><em>Primary structure</em>: the amino acid sequence </li> |
| − | <li><em | + | <li><em>Secondary structure</em>: regularly repeating local structures stabilized by hydrogen bonds. The most common examples are the alpha helix and beta sheet.<sup class="reference" id="_ref-Branden_0">[10]</sup> Because secondary structures are local, many regions of different secondary structure can be present in the same protein molecule. </li> |
| − | <li><em | + | <li><em>Tertiary structure</em>: the overall shape of a single protein molecule; the spatial relationship of the secondary structures to one another. Tertiary structure is generally stabilized by nonlocal interactions, most commonly the formation of a hydrophobic core, but also through salt bridges, hydrogen bonds, disulfide bonds, and even post-translational modifications. The term "tertiary structure" is often used as synonymous with the term <em>fold</em>. </li> |
| − | <li><em | + | <li><em>Quaternary structure</em>: the shape or structure that results from the interaction of more than one protein molecule, usually called <em>protein subunits</em> in this context, which function as part of the larger assembly or protein complex. </li> |
</ul> | </ul> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 152px | + | <div class="thumbinner" style="WIDTH: 152px"><img class="thumbimage" height="122" alt="NMR structures of the protein cytochrome c in solution show the constantly shifting dynamic structure of the protein. Larger version." width="150" longdesc="/wiki/Image:Protein_Dynamics_Cytochrome_C_2NEW_smaller.gif" src="http://upload.wikimedia.org/wikipedia/en/a/ae/Protein_Dynamics_Cytochrome_C_2NEW_smaller.gif" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | NMR structures of the protein | + | NMR structures of the protein cytochrome c in solution show the constantly shifting dynamic structure of the protein. Larger version.</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>Proteins are not entirely rigid molecules. In addition to these levels of structure, proteins may shift between several related structures in performing their biological function. In the context of these functional rearrangements, these tertiary or quaternary structures are usually referred to as " | + | <p>Proteins are not entirely rigid molecules. In addition to these levels of structure, proteins may shift between several related structures in performing their biological function. In the context of these functional rearrangements, these tertiary or quaternary structures are usually referred to as "conformations," and transitions between them are called <em>conformational changes.</em> Such changes are often induced by the binding of a substrate molecule to an enzyme's active site, or the physical region of the protein that participates in chemical catalysis. In solution all proteins also undergo variation in structure through thermal vibration and the collision with other molecules, see the animation on the right.</p> |
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 502px | + | <div class="thumbinner" style="WIDTH: 502px"><img class="thumbimage" height="144" alt="Molecular surface of several proteins showing their comparative sizes. From left to right are: Antibody (IgG), Hemoglobin, Insulin (a hormone), Adenylate kinase (an enzyme), and Glutamine synthetase (an enzyme)." width="500" longdesc="/wiki/Image:Protein_Composite.jpg" src="http://upload.wikimedia.org/wikipedia/commons/thumb/5/53/Protein_Composite.jpg/500px-Protein_Composite.jpg" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | Molecular surface of several proteins showing their comparative sizes. From left to right are: | + | Molecular surface of several proteins showing their comparative sizes. From left to right are: Antibody (IgG), Hemoglobin, Insulin (a hormone), Adenylate kinase (an enzyme), and Glutamine synthetase (an enzyme).</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: | + | <p>Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural; membrane proteins often serve as receptors or provide channels for polar or charged molecules to pass through the cell membrane.</p> |
| − | <p>A special case of intramolecular hydrogen bonds within proteins, poorly shielded from water attack and hence promoting their own | + | <p>A special case of intramolecular hydrogen bonds within proteins, poorly shielded from water attack and hence promoting their own dehydration, are called dehydrons.</p> |
<p><a id="Structure_determination" name="Structure_determination"></a></p> | <p><a id="Structure_determination" name="Structure_determination"></a></p> | ||
<h3><span class="mw-headline">Structure determination</span></h3> | <h3><span class="mw-headline">Structure determination</span></h3> | ||
| − | <p>Discovering the tertiary structure of a protein, or the quaternary structure of its complexes, can provide important clues about how the protein performs its function. Common experimental methods of structure determination include | + | <p>Discovering the tertiary structure of a protein, or the quaternary structure of its complexes, can provide important clues about how the protein performs its function. Common experimental methods of structure determination include X-ray crystallography and NMR spectroscopy, both of which can produce information at atomic resolution. Cryoelectron microscopy is used to produce lower-resolution structural information about very large protein complexes, including assembled viruses;<sup class="reference" id="_ref-Branden_1">[10]</sup> a variant known as electron crystallography can also produce high-resolution information in some cases, especially for two-dimensional crystals of membrane proteins.<sup class="reference" id="_ref-7">[11]</sup> Solved structures are usually deposited in the Protein Data Bank (PDB), a freely available resource from which structural data about thousands of proteins can be obtained in the form of Cartesian coordinates for each atom in the protein.</p> |
| − | <p>There are many more known gene sequences than there are solved protein structures. Further, the set of solved structures is biased toward those proteins that can be easily subjected to the experimental conditions required by one of the major structure determination methods. In particular, globular proteins are comparatively easy to | + | <p>There are many more known gene sequences than there are solved protein structures. Further, the set of solved structures is biased toward those proteins that can be easily subjected to the experimental conditions required by one of the major structure determination methods. In particular, globular proteins are comparatively easy to crystallize in preparation for X-ray crystallography, which remains the oldest and most common structure determination technique. Membrane proteins, by contrast, are difficult to crystallize and are underrepresented in the PDB.<sup class="reference" id="_ref-8">[12]</sup> Structural genomics initiatives have attempted to remedy these deficiencies by systematically solving representative structures of major fold classes. Protein structure prediction methods attempt to provide a means of generating a plausible structure for proteins whose structures have not been experimentally determined.</p> |
<p><a id="Cellular_functions" name="Cellular_functions"></a></p> | <p><a id="Cellular_functions" name="Cellular_functions"></a></p> | ||
<h2><span class="mw-headline">Cellular functions</span></h2> | <h2><span class="mw-headline">Cellular functions</span></h2> | ||
| − | <p>Proteins are the chief actors within the cell, said to be carrying out the duties specified by the information encoded in genes.<sup class="reference" id="_ref-Lodish_2 | + | <p>Proteins are the chief actors within the cell, said to be carrying out the duties specified by the information encoded in genes.<sup class="reference" id="_ref-Lodish_2">[5]</sup> With the exception of certain types of RNA, most other biological molecules are relatively inert elements upon which proteins act. Proteins make up half the dry weight of an <em>Escherichia coli</em> cell, while other macromolecules such as DNA and RNA make up only 3% and 20% respectively.<sup class="reference" id="_ref-Voet_0">[13]</sup> The set of proteins expressed in a particular cell or cell type is known as its proteome.</p> |
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 352px | + | <div class="thumbinner" style="WIDTH: 352px"><img class="thumbimage" height="250" alt="The enzyme hexokinase is shown as a simple ball-and-stick molecular model. To scale in the top right-hand corner are its two substrates, ATP and glucose." width="350" longdesc="/wiki/Image:Hexokinase_ball_and_stick_model%2C_with_substrates_to_scale_copy.png" src="http://upload.wikimedia.org/wikipedia/en/thumb/e/e7/Hexokinase_ball_and_stick_model%2C_with_substrates_to_scale_copy.png/350px-Hexokinase_ball_and_stick_model%2C_with_substrates_to_scale_copy.png" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | The enzyme | + | The enzyme hexokinase is shown as a simple ball-and-stick molecular model. To scale in the top right-hand corner are its two substrates, ATP and glucose.</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>The chief characteristic of proteins that enables them to carry out their diverse cellular functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the | + | <p>The chief characteristic of proteins that enables them to carry out their diverse cellular functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar dissociation constant (<10<sup>-15</sup> M) but does not bind at all to its amphibian homolog onconase (>1 M). Extremely minor chemical changes such as the addition of a single methyl group to a binding partner can sometimes suffice to nearly eliminate binding; for example, the aminoacyl tRNA synthetase specific to the amino acid valine discriminates against the very similar side chain of the amino acid isoleucine.</p> |
| − | <p>Proteins can bind to other proteins as well as to small-molecule substrates. When proteins bind specifically to other copies of the same molecule, they can | + | <p>Proteins can bind to other proteins as well as to small-molecule substrates. When proteins bind specifically to other copies of the same molecule, they can oligomerize to form fibrils; this process occurs often in structural proteins that consist of globular monomers that self-associate to form rigid fibers. Protein-protein interactions also regulate enzymatic activity, control progression through the cell cycle, and allow the assembly of large protein complexes that carry out many closely related reactions with a common biological function. Proteins can also bind to, or even be integrated into, cell membranes. The ability of binding partners to induce conformational changes in proteins allows the construction of enormously complex signaling networks.</p> |
<p><a id="Enzymes" name="Enzymes"></a></p> | <p><a id="Enzymes" name="Enzymes"></a></p> | ||
<h3><span class="mw-headline">Enzymes</span></h3> | <h3><span class="mw-headline">Enzymes</span></h3> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main article: | + | <div class="noprint"><em>Main article: Enzyme</em></div> |
</dd></dl> | </dd></dl> | ||
| − | <p>The best-known role of proteins in the cell is their duty as | + | <p>The best-known role of proteins in the cell is their duty as enzymes, which catalyze chemical reactions. Enzymes are usually highly specific catalysts that accelerate only one or a few chemical reactions. Enzymes effect most of the reactions involved in metabolism and catabolism as well as DNA replication, DNA repair, and RNA synthesis. Some enzymes act on other proteins to add or remove chemical groups in a process known as post-translational modification. About 4,000 reactions are known to be catalyzed by enzymes.<sup class="reference" id="_ref-9">[14]</sup> The rate acceleration conferred by enzymatic catalysis is often enormous - as much as 10<sup>17</sup>-fold increase in rate over the uncatalyzed reaction in the case of orotate decarboxylase (78 million years without the enzyme, 18 milliseconds with the enzyme).<sup class="reference" id="_ref-10">[15]</sup></p> |
| − | <p>The molecules bound and acted upon by enzymes are known as | + | <p>The molecules bound and acted upon by enzymes are known as substrates. Although enzymes can consist of hundreds of amino acids, it is usually only a small fraction of the residues that come in contact with the substrate and an even smaller fraction - 3-4 residues on average - that are directly involved in catalysis.<sup class="reference" id="_ref-11">[16]</sup> The region of the enzyme that binds the substrate and contains the catalytic residues is known as the active site.</p> |
<p><a id="Cell_signalling_and_ligand_transport" name="Cell_signalling_and_ligand_transport"></a></p> | <p><a id="Cell_signalling_and_ligand_transport" name="Cell_signalling_and_ligand_transport"></a></p> | ||
<h3><span class="mw-headline">Cell signalling and ligand transport</span></h3> | <h3><span class="mw-headline">Cell signalling and ligand transport</span></h3> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 152px | + | <div class="thumbinner" style="WIDTH: 152px"><img class="thumbimage" height="224" alt="A mouse antibody against cholera that binds a carbohydrate antigen." width="150" longdesc="/wiki/Image:Mouse-cholera-antibody-1f4x.png" src="http://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Mouse-cholera-antibody-1f4x.png/150px-Mouse-cholera-antibody-1f4x.png" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | A | + | A mouse antibody against cholera that binds a carbohydrate antigen.</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>Many proteins are involved in the process of | + | <p>Many proteins are involved in the process of cell signaling and signal transduction. Some proteins, such as insulin, are extracellular proteins that transmit a signal from the cell in which they were synthesized to other cells in distant tissues. Others are membrane proteins that act as receptors whose main function is to bind a signaling molecule and induce a biochemical response in the cell. Many receptors have a binding site exposed on the cell surface and an effector domain within the cell, which may have enzymatic activity or may undergo a conformational change detected by other proteins within the cell.</p> |
| − | <p | + | <p>Antibodies are protein components of adaptive immune system whose main function is to bind antigens, or foreign substances in the body, and target them for destruction. Antibodies can be secreted into the extracellular environment or anchored in the membranes of specialized B cells known as plasma cells. While enzymes are limited in their binding affinity for their substrates by the necessity of conducting their reaction, antibodies have no such constraints. An antibody's binding affinity to its target is extraordinarily high.</p> |
| − | <p>Many ligand transport proteins bind particular small biomolecules and transport them to other locations in the body of a multicellular organism. These proteins must have a high binding affinity when their | + | <p>Many ligand transport proteins bind particular small biomolecules and transport them to other locations in the body of a multicellular organism. These proteins must have a high binding affinity when their ligand is present in high concentrations but must also release the ligand when it is present at low concentrations in the target tissues. The canonical example of a ligand-binding protein is haemoglobin, which transports oxygen from the lungs to other organs and tissues in all vertebrates and has close homologs in every biological kingdom.</p> |
| − | <p | + | <p>Transmembrane proteins can also serve as ligand transport proteins that alter the permeability of the cell's membrane to small molecules and ions. The membrane alone has a hydrophobic core through which polar or charged molecules cannot diffuse. Membrane proteins contain internal channels that allow such molecules to enter and exit the cell. Many ion channel proteins are specialized to select for only a particular ion; for example, potassium and sodium channels often discriminate for only one of the two ions.</p> |
<p><a id="Structural_proteins" name="Structural_proteins"></a></p> | <p><a id="Structural_proteins" name="Structural_proteins"></a></p> | ||
<h3><span class="mw-headline">Structural proteins</span></h3> | <h3><span class="mw-headline">Structural proteins</span></h3> | ||
| − | <p>Structural proteins confer stiffness and rigidity to otherwise fluid biological components. Most structural proteins are | + | <p>Structural proteins confer stiffness and rigidity to otherwise fluid biological components. Most structural proteins are fibrous proteins; for example, actin and tubulin are globular and soluble as monomers but polymerize to form long, stiff fibers that comprise the cytoskeleton, which allows the cell to maintain its shape and size. Collagen and elastin are critical components of connective tissue such as cartilage, and keratin is found in hard or filamentous structures such as hair, nails, feathers, hooves, and some animal shells.</p> |
| − | <p>Other proteins that serve structural functions are | + | <p>Other proteins that serve structural functions are motor proteins such as myosin, kinesin, and dynein, which are capable of generating mechanical forces. These proteins are crucial for cellular motility of single-celled organisms and the sperm of many sexually reproducing multicellular organisms. They also generate the forces exerted by contracting muscles.</p> |
<p><a id="Methods_of_study" name="Methods_of_study"></a></p> | <p><a id="Methods_of_study" name="Methods_of_study"></a></p> | ||
<h2><span class="mw-headline">Methods of study</span></h2> | <h2><span class="mw-headline">Methods of study</span></h2> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main article: | + | <div class="noprint"><em>Main article: Protein methods</em></div> |
</dd></dl> | </dd></dl> | ||
| − | <p>As some of the most commonly studied biological molecules, the activities and structures of proteins are examined both <em>in vitro</em> and <em>in vivo</em>. <em>In vitro</em> studies of purified proteins in controlled environments are useful for learning how a protein carries out its function: for example, | + | <p>As some of the most commonly studied biological molecules, the activities and structures of proteins are examined both <em>in vitro</em> and <em>in vivo</em>. <em>In vitro</em> studies of purified proteins in controlled environments are useful for learning how a protein carries out its function: for example, enzyme kinetics studies explore the chemical mechanism of an enzyme's catalytic activity and its relative affinity for various possible substrate molecules. By contrast, <em>in vivo</em> experiments on proteins' activities within cells or even within whole organisms can provide complementary information about where a protein functions and how it is regulated.</p> |
<p><a id="Protein_purification" name="Protein_purification"></a></p> | <p><a id="Protein_purification" name="Protein_purification"></a></p> | ||
<h3><span class="mw-headline">Protein purification</span></h3> | <h3><span class="mw-headline">Protein purification</span></h3> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main article: | + | <div class="noprint"><em>Main article: Protein purification</em></div> |
</dd></dl> | </dd></dl> | ||
| − | <p>In order to perform <em>in vitro</em> analyses, a protein must be purified away from other cellular components. This process usually begins with | + | <p>In order to perform <em>in vitro</em> analyses, a protein must be purified away from other cellular components. This process usually begins with cell lysis, in which a cell's membrane is disrupted and its internal contents released into a solution known as a crude lysate. The resulting mixture can be purified using ultracentrifugation, which fractionates the various cellular components into fractions containing soluble proteins; membrane lipids and proteins; cellular organelles, and nucleic acids. Precipitation by a method known as salting out can concentrate the proteins from this lysate. Various types of chromatography are then used to isolate the protein or proteins of interest based on properties such as molecular weight, net charge and binding affinity. The level of purification can be monitored using gel electrophoresis if the desired protein's molecular weight is known, by spectroscopy if the protein has distinguishable spectroscopic features, or by enzyme assays if the protein has enzymatic activity.</p> |
| − | <p>For natural proteins, a series of purification steps may be necessary to obtain protein sufficiently pure for laboratory applications. To simplify this process, | + | <p>For natural proteins, a series of purification steps may be necessary to obtain protein sufficiently pure for laboratory applications. To simplify this process, genetic engineering is often used to add chemical features to proteins that make them easier to purify without affecting their structure or activity. Here, a "tag" consisting of a specific amino acid sequence, often a series of histidine residues (a "His-tag"), is attached to one terminus of the protein. As a result, when the lysate is passed over a chromatography column containing nickel, the histidine residues ligate the nickel and attach to the column while the untagged components of the lysate pass unimpeded.</p> |
<p><a id="Cellular_localization" name="Cellular_localization"></a></p> | <p><a id="Cellular_localization" name="Cellular_localization"></a></p> | ||
<h3><span class="mw-headline">Cellular localization</span></h3> | <h3><span class="mw-headline">Cellular localization</span></h3> | ||
<div class="thumb tright"> | <div class="thumb tright"> | ||
| − | <div class="thumbinner" style="WIDTH: 302px | + | <div class="thumbinner" style="WIDTH: 302px"><img class="thumbimage" height="395" alt="Proteins in different cellular compartments and structures tagged with green fluorescent protein (here, white)." width="300" longdesc="/wiki/Image:Localisations02eng.jpg" src="http://upload.wikimedia.org/wikipedia/commons/thumb/6/6e/Localisations02eng.jpg/300px-Localisations02eng.jpg" /> |
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| − | <div class="magnify" style="FLOAT: right | + | <div class="magnify" style="FLOAT: right"><img height="11" alt="" width="15" src="http://en.wikipedia.org/skins-1.5/common/images/magnify-clip.png" /></div> |
| − | Proteins in different | + | Proteins in different cellular compartments and structures tagged with green fluorescent protein (here, white).</div> |
</div> | </div> | ||
</div> | </div> | ||
| − | <p>The study of proteins <em>in vivo</em> is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the | + | <p>The study of proteins <em>in vivo</em> is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the endoplasmic reticulum, the specifics of how proteins are targeted to specific organelles or cellular structures is often unclear. A useful technique for assessing cellular localization uses genetic engineering to express in a cell a fusion protein or chimera consisting of the natural protein of interest linked to a "reporter" such as green fluorescent protein (GFP). The fused protein's position within the cell can be cleanly and efficiently visualized using microscopy, as shown in the figure opposite.</p> |
| − | <p>Through another genetic engineering application known as | + | <p>Through another genetic engineering application known as site-directed mutagenesis, researchers can alter the protein sequence and hence its structure, cellular localization, and susceptibility to regulation, which can be followed <em>in vivo</em> by GFP tagging or <em>in vitro</em> by enzyme kinetics and binding studies.</p> |
<p><a id="Proteomics_and_bioinformatics" name="Proteomics_and_bioinformatics"></a></p> | <p><a id="Proteomics_and_bioinformatics" name="Proteomics_and_bioinformatics"></a></p> | ||
<h3><span class="mw-headline">Proteomics and bioinformatics</span></h3> | <h3><span class="mw-headline">Proteomics and bioinformatics</span></h3> | ||
<dl><dd> | <dl><dd> | ||
| − | <div class="noprint"><em>Main articles: | + | <div class="noprint"><em>Main articles: Proteomics and Bioinformatics</em></div> |
</dd></dl> | </dd></dl> | ||
| − | <p>The total complement of proteins present at a time in a cell or cell type is known as its | + | <p>The total complement of proteins present at a time in a cell or cell type is known as its proteome, and the study of such large-scale data sets defines the field of proteomics, named by analogy to the related field of genomics. Key experimental techniques in proteomics include protein microarrays, which allow the detection of the relative levels of a large number of proteins present in a cell, and two-hybrid screening, which allows the systematic exploration of protein-protein interactions. The total complement of biologically possible such interactions is known as the interactome. A systematic attempt to determine the structures of proteins representing every possible fold is known as structural genomics.</p> |
| − | <p>The large amount of genomic and proteomic data available for a variety of organisms, including the | + | <p>The large amount of genomic and proteomic data available for a variety of organisms, including the human genome, allows researchers to efficiently identify homologous proteins in distantly related organisms by sequence alignment. Sequence profiling tools can perform more specific sequence manipulations such as restriction enzyme maps, open reading frame analyses for nucleotide sequences, and secondary structure prediction. From this data phylogenetic trees can be constructed and evolutionary hypotheses developed using special software like ClustalW regarding the ancestry of modern organisms and the genes they express. The field of bioinformatics seeks to assemble, annotate, and analyze genomic and proteomic data, applying computational techniques to biological problems such as gene finding and cladistics.</p> |
<p><a id="Structure_prediction_and_simulation" name="Structure_prediction_and_simulation"></a></p> | <p><a id="Structure_prediction_and_simulation" name="Structure_prediction_and_simulation"></a></p> | ||
<h3><span class="mw-headline">Structure prediction and simulation</span></h3> | <h3><span class="mw-headline">Structure prediction and simulation</span></h3> | ||
| − | <p>Complementary to the field of structural genomics, | + | <p>Complementary to the field of structural genomics, protein structure prediction seeks to develop efficient ways to provide plausible models for proteins whose structures have not yet been determined experimentally. The most successful type of structure prediction, known as homology modeling, relies on the existence of a "template" structure with sequence similarity to the protein being modeled; structural genomics' goal is to provide sufficient representation in solved structures to model most of those that remain. Although producing accurate models remains a challenge when only distantly related template structures are available, it has been suggested that sequence alignment is the bottleneck in this process, as quite accurate models can be produced if a "perfect" sequence alignment is known.<sup class="reference" id="_ref-Zhang_0">[17]</sup> Many structure prediction methods have served to inform the emerging field of protein engineering, in which novel protein folds have already been designed.<sup class="reference" id="_ref-Kuhlman_0">[18]</sup> A more complex computational problem is the prediction of intermolecular interactions, such as in molecular docking and protein-protein interaction prediction.</p> |
| − | <p>The processes of protein folding and binding can be simulated using techniques derived from | + | <p>The processes of protein folding and binding can be simulated using techniques derived from molecular dynamics, which increasingly take advantage of distributed computing as in the Folding@Home project. The folding of small alpha-helical protein domains such as the villin headpiece<sup class="reference" id="_ref-Zagrovic_0">[19]</sup> and the HIV accessory protein<sup class="reference" id="_ref-Herges_0">[20]</sup> have been successfully simulated <em>in silico</em>, and hybrid methods that combine standard molecular dynamics with quantum mechanics calculations have allowed exploration of the electronic states of rhodopsins.<sup class="reference" id="_ref-Hoffmann_0">[21]</sup></p> |
<p><a id="Visualization" name="Visualization"></a></p> | <p><a id="Visualization" name="Visualization"></a></p> | ||
<h2><span class="mw-headline">Visualization</span></h2> | <h2><span class="mw-headline">Visualization</span></h2> | ||
<ul> | <ul> | ||
| − | <li> | + | <li>PyMOL </li> |
| − | <li | + | <li>Sirius </li> |
| − | <li | + | <li>Rasmol </li> |
| − | <li | + | <li>Visual molecular dynamics </li> |
</ul> | </ul> | ||
<p><a id="Nutrition" name="Nutrition"></a></p> | <p><a id="Nutrition" name="Nutrition"></a></p> | ||
<h2><span class="mw-headline">Nutrition</span></h2> | <h2><span class="mw-headline">Nutrition</span></h2> | ||
| − | <dl><dd><em>Further information: | + | <dl><dd><em>Further information: Protein in nutrition</em> </dd></dl> |
| − | <p>Most | + | <p>Most microorganisms and plants can biosynthesize all 20 standard amino acids, while animals must obtain some of the amino acids from the diet.<sup class="reference" id="_ref-Voet_1">[13]</sup> Key enzymes in the biosynthetic pathways that synthesize certain amino acids - such as aspartokinase, which catalyzes the first step in the synthesis of lysine, methionine, and threonine from aspartate - are not present in animals. The amino acids that an organism cannot synthesize on its own are referred to as essential amino acids. (This designation is often used to specifically identify those essential to humans.) If amino acids are present in the environment, most microorganisms can conserve energy by taking up the amino acids from the environment and downregulating their own biosynthetic pathways. Bacteria are often engineered in the laboratory to lack the genes necessary for synthesizing a particular amino acid, providing a selectable marker for the success of transfection, or the introduction of foreign DNA.</p> |
| − | <p>In animals, amino acids are obtained through the consumption of foods containing protein. Ingested proteins are broken down through | + | <p>In animals, amino acids are obtained through the consumption of foods containing protein. Ingested proteins are broken down through digestion, which typically involves denaturation of the protein through exposure to acid and degradation by the action of enzymes called proteases. Ingestion of essential amino acids is critical to the health of the organism, since the biosynthesis of proteins that include these amino acids is inhibited by their low concentration. Amino acids are also an important dietary source of nitrogen. Some ingested amino acids, especially those that are not essential, are not used directly for protein biosynthesis. Instead, they are converted to carbohydrates through gluconeogenesis, which is also used under starvation conditions to generate glucose from the body's own proteins, particularly those found in muscle.</p> |
<p><a id="History" name="History"></a></p> | <p><a id="History" name="History"></a></p> | ||
<h2><span class="mw-headline">History</span></h2> | <h2><span class="mw-headline">History</span></h2> | ||
| − | <dl><dd><em>Further information: | + | <dl><dd><em>Further information: History of molecular biology</em> </dd></dl> |
| − | <p>Proteins were recognized as a distinct class of biological molecules in the eighteenth century by | + | <p>Proteins were recognized as a distinct class of biological molecules in the eighteenth century by Antoine Fourcroy and others, distinguished by the molecules' ability to coagulate or flocculate under treatments with heat or acid. Noted examples at the time included albumen from egg whites, blood, serum albumin, fibrin, and wheat gluten. Dutch chemist Gerhardus Johannes Mulder carried out elemental analysis of common proteins and found that nearly all proteins had the same empirical formula. The term "protein" to describe these molecules was proposed in 1838 by Mulder's associate Jöns Jakob Berzelius. Mulder went on to identify the products of protein degradation such as the amino acid leucine for which he found a (nearly correct) molecular weight of 131 Da.</p> |
| − | <p>The difficulty in purifying proteins in large quantities made them very difficult for early protein biochemists to study. Hence, early studies focused on proteins that could be purified in large quantities, e.g., those of | + | <p>The difficulty in purifying proteins in large quantities made them very difficult for early protein biochemists to study. Hence, early studies focused on proteins that could be purified in large quantities, e.g., those of blood, egg white, various toxins, and digestive/metabolic enzymes obtained from slaughterhouses. In the late 1950s, the Armour Hot Dog Co. purified 1 kg (= one million milligrams) of pure bovine pancreatic ribonuclease A and made it freely available to scientists around the world.</p> |
| − | <p> | + | <p>Linus Pauling is credited with the successful prediction of regular protein secondary structures based on hydrogen bonding, an idea first put forth by William Astbury in 1933. Later work by Walter Kauzman on denaturation, based partly on previous studies by Kaj Linderstrom-Lang, contributed an understanding of protein folding and structure mediated by hydrophobic interactions. In 1949 Fred Sanger correctly determined the amino acid sequence of insulin, thus conclusively demonstrating that proteins consisted of linear polymers of amino acids rather than branched chains, colloids, or cyclols. The first atomic-resolution structures of proteins were solved by X-ray crystallography in the 1960s and by NMR in the 1980s. As of 2006, the Protein Data Bank has nearly 40,000 atomic-resolution structures of proteins. In more recent times, cryo-electron microscopy of large macromolecular assemblies and computational protein structure prediction of small protein domains are two methods approaching atomic resolution.</p> |
<p><a id="See_also" name="See_also"></a></p> | <p><a id="See_also" name="See_also"></a></p> | ||
<h2><span class="mw-headline">See also</span></h2> | <h2><span class="mw-headline">See also</span></h2> | ||
<ul> | <ul> | ||
| − | <li | + | <li>Amino acid </li> |
| − | <li | + | <li>Essential amino acid </li> |
| − | <li | + | <li>Protein design </li> |
| − | <li> | + | <li>Intein </li> |
| − | <li | + | <li>List of recombinant proteins </li> |
| − | <li | + | <li>List of proteins </li> |
| − | <li> | + | <li>Prion </li> |
| − | <li | + | <li>Edible protein per unit area of land </li> |
| − | <li> | + | <li>Expression cloning </li> |
</ul> | </ul> | ||
<p><a id="References" name="References"></a></p> | <p><a id="References" name="References"></a></p> | ||
| Line 184: | Line 183: | ||
<div class="references-2column"> | <div class="references-2column"> | ||
<ol class="references"> | <ol class="references"> | ||
| − | <li id="_note-0"><strong | + | <li id="_note-0"><strong>^</strong> <cite style="FONT-STYLE: normal">Sumner, JB (1926). "The Isolation and Crystallization of the Enzyme Urease. Preliminary Paper". <em>J Biol Chem</em> <strong>69</strong>: 435-41.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=The+Isolation+and+Crystallization+of+the+Enzyme+Urease.+Preliminary+Paper&rft.jtitle=J+Biol+Chem&rft.date=1926&rft.volume=69&rft.au=Sumner%2C+JB&rft.pages=435-41&rft_id=http%3A%2F%2Fwww.jbc.org%2Fcgi%2Freprint%2F69%2F2%2F435.pdf%3Fijkey%3D028d5e540dab50accbf86e01be08db51ef49008f"> </span> </li> |
| − | + | <li id="_note-1"><strong>^</strong> <cite style="FONT-STYLE: normal">Muirhead H, Perutz M (1963). "Structure of haemoglobin. A three-dimensional fourier synthesis of reduced human haemoglobin at 5.5 A resolution". <em>Nature</em> <strong>199</strong> (4894): 633-8. PMID 14074546.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Structure+of+haemoglobin.+A+three-dimensional+fourier+synthesis+of+reduced+human+haemoglobin+at+5.5+A+resolution&rft.jtitle=Nature&rft.date=1963&rft.volume=199&rft.issue=4894&rft.au=Muirhead+H%2C+Perutz+M&rft.pages=633-8&rft_id=info:pmid/14074546"> </span> </li> | |
| − | + | <li id="_note-2"><strong>^</strong> <cite style="FONT-STYLE: normal">Kendrew J, Bodo G, Dintzis H, Parrish R, Wyckoff H, Phillips D (1958). "A three-dimensional model of the myoglobin molecule obtained by x-ray analysis". <em>Nature</em> <strong>181</strong> (4610): 662-6. PMID 13517261.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=A+three-dimensional+model+of+the+myoglobin+molecule+obtained+by+x-ray+analysis&rft.jtitle=Nature&rft.date=1958&rft.volume=181&rft.issue=4610&rft.au=Kendrew+J%2C+Bodo+G%2C+Dintzis+H%2C+Parrish+R%2C+Wyckoff+H%2C+Phillips+D&rft.pages=662-6&rft_id=info:pmid/13517261"> </span> </li> | |
| − | + | <li id="_note-3"><strong>^</strong> Nelson, D. L. and Cox, M. M. (2005) Lehninger's Principles of Biochemistry, 4th Edition, W. H. Freeman and Company, New York. </li> | |
| − | + | <li id="_note-Lodish">^ <sup><em><strong>a</strong></em></sup> <sup><em><strong>b</strong></em></sup> <sup><em><strong>c</strong></em></sup> Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipurksy SL, Darnell J. (2004). <em>Molecular Cell Biology</em> 5th ed. WH Freeman and Company: New York, NY. </li> | |
| − | + | <li id="_note-Dobson"><strong>^</strong> Dobson CM. (2000). The nature and significance of protein folding. In <em>Mechanisms of Protein Folding</em> 2nd ed. Ed. RH Pain. <em>Frontiers in Molecular Biology</em> series. Oxford University Press: New York, NY. </li> | |
| − | + | <li id="_note-4"><strong>^</strong> <cite style="FONT-STYLE: normal">Fulton A, Isaacs W (1991). "Titin, a huge, elastic sarcomeric protein with a probable role in morphogenesis". <em>Bioessays</em> <strong>13</strong> (4): 157-61. PMID 1859393.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Titin%2C+a+huge%2C+elastic+sarcomeric+protein+with+a+probable+role+in+morphogenesis&rft.jtitle=Bioessays&rft.date=1991&rft.volume=13&rft.issue=4&rft.au=Fulton+A%2C+Isaacs+W&rft.pages=157-61&rft_id=info:pmid/1859393"> </span> </li> | |
| − | + | <li id="_note-5"><strong>^</strong> <cite style="FONT-STYLE: normal">Bruckdorfer T, Marder O, Albericio F (2004). "From production of peptides in milligram amounts for research to multi-tons quantities for drugs of the future". <em>Curr Pharm Biotechnol</em> <strong>5</strong> (1): 29-43. PMID 14965208.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=From+production+of+peptides+in+milligram+amounts+for+research+to+multi-tons+quantities+for+drugs+of+the+future&rft.jtitle=Curr+Pharm+Biotechnol&rft.date=2004&rft.volume=5&rft.issue=1&rft.au=Bruckdorfer+T%2C+Marder+O%2C+Albericio+F&rft.pages=29-43&rft_id=info:pmid/14965208"> </span> </li> | |
| − | + | <li id="_note-6"><strong>^</strong> <cite style="FONT-STYLE: normal">Schwarzer D, Cole P (2005). "Protein semisynthesis and expressed protein ligation: chasing a protein's tail". <em>Curr Opin Chem Biol</em> <strong>9</strong> (6): 561-9. PMID 16226484.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=Protein+semisynthesis+and+expressed+protein+ligation%3A+chasing+a+protein%27s+tail&rft.jtitle=Curr+Opin+Chem+Biol&rft.date=2005&rft.volume=9&rft.issue=6&rft.au=Schwarzer+D%2C+Cole+P&rft.pages=561-9&rft_id=info:pmid/16226484"> </span> </li> | |
| − | + | <li id="_note-Branden">^ <sup><em><strong>a</strong></em></sup> <sup><em><strong>b</strong></em></sup> Branden C, Tooze J. (1999). <em>Introduction to Protein Structure</em> 2nd ed. Garland Publishing: New York, NY </li> | |
| − | + | <li id="_note-7"><strong>^</strong> Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. (2005). Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. <em>Nature</em> 438(7068):633-8. </li> | |
| − | + | <li id="_note-8"><strong>^</strong> Walian P, Cross TA, Jap BK. (2004). Structural genomics of membrane proteins <em>Genome Biol</em> 5(4): 215. </li> | |
| − | + | <li id="_note-Voet">^ <sup><em><strong>a</strong></em></sup> <sup><em><strong>b</strong></em></sup> Voet D, Voet JG. (2004). <em>Biochemistry</em> Vol 1 3rd ed. Wiley: Hoboken, NJ. </li> | |
| − | + | <li id="_note-9"><strong>^</strong> <cite style="FONT-STYLE: normal">Bairoch A. (2000). "The ENZYME database in 2000". <em>Nucleic Acids Res</em> <strong>28</strong>: 304-305. PMID 10592255.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=The+ENZYME+database+in+2000&rft.jtitle=Nucleic+Acids+Res&rft.date=2000&rft.volume=28&rft.au=Bairoch+A.&rft.pages=304-305&rft_id=http%3A%2F%2Fwww.expasy.org%2FNAR%2Fenz00.pdf"> </span> </li> | |
| − | <li id="_note-10"><strong | + | <li id="_note-10"><strong>^</strong> <cite style="FONT-STYLE: normal">Radzicka A, Wolfenden R. (1995). "A proficient enzyme.". <em>Science</em> <strong>6</strong> (267): 90-931. PMID 7809611.</cite><span class="Z3988" title="ctx_ver=Z39.88-2004&rft_val_fmt=info%3Aofi%2Ffmt%3Akev%3Amtx%3Ajournal&rft.genre=article&rft.atitle=A+proficient+enzyme.&rft.jtitle=Science&rft.date=1995&rft.volume=6&rft.issue=267&rft.au=Radzicka+A%2C+Wolfenden+R.&rft.pages=90-931"> </span> </li> |
| − | <li id="_note-11"><strong | + | <li id="_note-11"><strong>^</strong> The Catalytic Site Atlas at The European Bioinformatics Institute </li> |
| − | <li id="_note-Zhang"><strong | + | <li id="_note-Zhang"><strong>^</strong> Zhang Y, Skolnick J. (2005). The protein structure prediction problem could be solved using the current PDB library. <em>Proc Natl Acad Sci USA</em> 102(4):1029-34. </li> |
| − | <li id="_note-Kuhlman"><strong | + | <li id="_note-Kuhlman"><strong>^</strong> Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. (2003). Design of a novel globular protein fold with atomic-level accuracy. <em>Science</em> 302(5649):1364-8. </li> |
| − | <li id="_note-Zagrovic"><strong | + | <li id="_note-Zagrovic"><strong>^</strong> Zagrovic B, Snow CD, Shirts MR, Pande VS. (2002). Simulation of folding of a small alpha-helical protein in atomistic detail using worldwide-distributed computing. <em>J Mol Biol</em> 323(5):927-37. </li> |
| − | <li id="_note-Herges"><strong | + | <li id="_note-Herges"><strong>^</strong> Herges T, Wenzel W. (2005). In silico folding of a three helix protein and characterization of its free-energy landscape in an all-atom force field. <em>Phys Rev Let</em> 94(1):018101. </li> |
| − | <li id="_note-Hoffmann"><strong | + | <li id="_note-Hoffmann"><strong>^</strong> Hoffmann M, Wanko M, Strodel P, Konig PH, Frauenheim T, Schulten K, Thiel W, Tajkhorshid E, Elstner M. (2006). Color tuning in rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. <em>J Am Chem Soc</em> 128(33):10808-18. </li> |
</ol> | </ol> | ||
</div> | </div> | ||
| Line 210: | Line 209: | ||
<h2><span class="mw-headline">External links</span></h2> | <h2><span class="mw-headline">External links</span></h2> | ||
<ul> | <ul> | ||
| − | <li | + | <li>Proteins (the journal), also called "Proteins: Structure, Function, and Bioinformatics" and previously "Proteins: Structure, Function, and Genetics" (1986-1995). </li> |
</ul> | </ul> | ||
<p><a id="Databases_and_projects" name="Databases_and_projects"></a></p> | <p><a id="Databases_and_projects" name="Databases_and_projects"></a></p> | ||
Latest revision as of 13:48, 26 February 2008
Proteins are large organic compounds made of amino acids arranged in a linear chain and joined together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. The sequence of amino acids in a protein is defined by a gene and encoded in the genetic code. Although this genetic code specifies 20 "standard" amino acids, the residues in a protein are often chemically altered in post-translational modification: either before the protein can function in the cell, or as part of control mechanisms. Proteins can also work together to achieve a particular function, and they often associate to form stable complexes.
Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of all living organisms and participate in every process within cells. Many proteins are enzymes that catalyze biochemical reactions, and are vital to metabolism. Other proteins have structural or mechanical functions, such as the proteins in the cytoskeleton, which forms a system of scaffolding that maintains cell shape. Proteins are also important in cell signaling, immune responses, cell adhesion, and the cell cycle. Protein is also a necessary component in our diet, since animals cannot synthesise all the amino acids and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that can be used for protein synthesis.
The word protein comes from the Greek πρώτα ("prota"), meaning "of primary importance" and these molecules were first described and named by Jöns Jakob Berzelius in 1838. However, proteins' central role in living organisms was not fully appreciated until 1926, when James B. Sumner showed that the enzyme urease was a protein.[1] The first protein to be sequenced was insulin, by Frederick Sanger, who won the Nobel Prize for this achievement in 1958. The first protein structures to be solved included haemoglobin and myoglobin, by Max Perutz and Sir John Cowdery Kendrew, respectively, in 1958.[2][3] Both proteins' three-dimensional structures were first determined by x-ray diffraction analysis; the structures of myoglobin and haemoglobin won the 1962 Nobel Prize in Chemistry for their discoverers.
Contents
Biochemistry


Proteins are linear polymers built from 20 different L-α-amino acids. All amino acids share common structural features including an α carbon to which an amino group, a carboxyl group, and a variable side chain are bonded. Only proline differs from this basic structure, as it contains an unusual ring to the N-end amine group, which forces the CO-NH amide moiety into a fixed conformation.[4] The side chains of the standard amino acids, detailed in the list of standard amino acids, have different chemical properties that produce proteins' three-dimensional structure and are therefore critical to protein function. The amino acids in a polypeptide chain are linked by peptide bonds formed in a dehydration reaction. Once linked in the protein chain, an individual amino acid is called a residue and the linked series of carbon, nitrogen, and oxygen atoms are known as the main chain or protein backbone. The peptide bond has two resonance forms that contribute some double bond character and inhibit rotation around its axis, so that the alpha carbons are roughly coplanar. The other two dihedral angles in the peptide bond determine the local shape assumed by the protein backbone.
Due to the chemical structure of the individual amino acids, the protein chain has directionality. The end of the protein with a free carboxyl group is known as the C-terminus or carboxy terminus, while the end with a free amino group is known as the N-terminus or amino terminus.
There is some ambiguity between the usage of the words protein, polypeptide, and peptide. Protein is generally used to refer to the complete biological molecule in a stable conformation, while peptide is generally reserved for a short amino acid oligomers often lacking a stable 3-dimensional structure. However, the boundary between the two is ill-defined and usually lies near 20-30 residues.[5] Polypeptide can refer to any single linear chain of amino acids, usually regardless of length, but often implies an absence of a single defined conformation.
Synthesis

Proteins are assembled from amino acids using information encoded in genes. Each protein has its own unique amino acid sequence that is specified by the nucleotide sequence of the gene encoding this protein. The genetic code is a set of three-nucleotide sets called codons and each three-nucleotide combination stands for an amino acid, for example AUG stands for methionine. Because DNA contains four nucleotides, the total number of possible codons is 64; hence, there is some redundancy in the genetic code and some amino acids are specified by more than one codon. Genes encoded in DNA are first transcribed into pre-messenger RNA (mRNA) by proteins such as RNA polymerase. Most organisms then process the pre-mRNA (also known as a primary transcript) using various forms of post-transcriptional modification to form the mature mRNA, which is then used as a template for protein synthesis by the ribosome. In prokaryotes the mRNA may either be used as soon as it is produced, or be bound by a ribosome after having moved away from the nucleoid. In contrast, eukaryotes make mRNA in the cell nucleus and then translocate it across the nuclear membrane into the cytoplasm, where protein synthesis then takes place. The rate of protein synthesis is higher in prokaryotes than eukaryotes and can reach up to 20 amino acids per second.[6]
The process of synthesizing a protein from an mRNA template is known as translation. The mRNA is loaded onto the ribosome and is read three nucleotides at a time by matching each codon to its base pairing anticodon located on a transfer RNA molecule, which carries the amino acid corresponding to the codon it recognizes. The enzyme aminoacyl tRNA synthetase "charges" the tRNA molecules with the correct amino acids. The growing polypeptide is often termed the nascent chain. Proteins are always biosynthesized from N-terminus to C-terminus.
The size of a synthesized protein can be measured by the number of amino acids it contains and by its total molecular mass, which is normally reported in units of daltons (synonymous with atomic mass units), or the derivative unit kilodalton (kDa). Yeast proteins are on average 466 amino acids long and 53 kDa in mass.[5] The largest known proteins are the titins, a component of the muscle sarcomere, with a molecular mass of almost 3,000 kDa and a total length of almost 27,000 amino acids.[7]
Chemical synthesis
Short proteins can also be synthesized chemically by a family of methods known as peptide synthesis, which rely on organic synthesis techniques such as chemical ligation to produce peptides in high yield.[8] Chemical synthesis allows for the introduction of non-natural amino acids into polypeptide chains, such as attachment of fluorescent probes to amino acid side chains.[9] These methods are useful in laboratory biochemistry and cell biology, though generally not for commercial applications. Chemical synthesis is inefficient for polypeptides longer than about 300 amino acids, and the synthesized proteins may not readily assume their native tertiary structure. Most chemical synthesis methods proceed from C-terminus to N-terminus, opposite the biological reaction.
Structure of proteins

Most proteins fold into unique 3-dimensional structures. The shape into which a protein naturally folds is known as its native state. Although many proteins can fold unassisted simply through the structural propensities of their component amino acids, others require the aid of molecular chaperones to efficiently fold to their native states. Biochemists often refer to four distinct aspects of a protein's structure:
- Primary structure: the amino acid sequence
- Secondary structure: regularly repeating local structures stabilized by hydrogen bonds. The most common examples are the alpha helix and beta sheet.[10] Because secondary structures are local, many regions of different secondary structure can be present in the same protein molecule.
- Tertiary structure: the overall shape of a single protein molecule; the spatial relationship of the secondary structures to one another. Tertiary structure is generally stabilized by nonlocal interactions, most commonly the formation of a hydrophobic core, but also through salt bridges, hydrogen bonds, disulfide bonds, and even post-translational modifications. The term "tertiary structure" is often used as synonymous with the term fold.
- Quaternary structure: the shape or structure that results from the interaction of more than one protein molecule, usually called protein subunits in this context, which function as part of the larger assembly or protein complex.

Proteins are not entirely rigid molecules. In addition to these levels of structure, proteins may shift between several related structures in performing their biological function. In the context of these functional rearrangements, these tertiary or quaternary structures are usually referred to as "conformations," and transitions between them are called conformational changes. Such changes are often induced by the binding of a substrate molecule to an enzyme's active site, or the physical region of the protein that participates in chemical catalysis. In solution all proteins also undergo variation in structure through thermal vibration and the collision with other molecules, see the animation on the right.

Proteins can be informally divided into three main classes, which correlate with typical tertiary structures: globular proteins, fibrous proteins, and membrane proteins. Almost all globular proteins are soluble and many are enzymes. Fibrous proteins are often structural; membrane proteins often serve as receptors or provide channels for polar or charged molecules to pass through the cell membrane.
A special case of intramolecular hydrogen bonds within proteins, poorly shielded from water attack and hence promoting their own dehydration, are called dehydrons.
Structure determination
Discovering the tertiary structure of a protein, or the quaternary structure of its complexes, can provide important clues about how the protein performs its function. Common experimental methods of structure determination include X-ray crystallography and NMR spectroscopy, both of which can produce information at atomic resolution. Cryoelectron microscopy is used to produce lower-resolution structural information about very large protein complexes, including assembled viruses;[10] a variant known as electron crystallography can also produce high-resolution information in some cases, especially for two-dimensional crystals of membrane proteins.[11] Solved structures are usually deposited in the Protein Data Bank (PDB), a freely available resource from which structural data about thousands of proteins can be obtained in the form of Cartesian coordinates for each atom in the protein.
There are many more known gene sequences than there are solved protein structures. Further, the set of solved structures is biased toward those proteins that can be easily subjected to the experimental conditions required by one of the major structure determination methods. In particular, globular proteins are comparatively easy to crystallize in preparation for X-ray crystallography, which remains the oldest and most common structure determination technique. Membrane proteins, by contrast, are difficult to crystallize and are underrepresented in the PDB.[12] Structural genomics initiatives have attempted to remedy these deficiencies by systematically solving representative structures of major fold classes. Protein structure prediction methods attempt to provide a means of generating a plausible structure for proteins whose structures have not been experimentally determined.
Cellular functions
Proteins are the chief actors within the cell, said to be carrying out the duties specified by the information encoded in genes.[5] With the exception of certain types of RNA, most other biological molecules are relatively inert elements upon which proteins act. Proteins make up half the dry weight of an Escherichia coli cell, while other macromolecules such as DNA and RNA make up only 3% and 20% respectively.[13] The set of proteins expressed in a particular cell or cell type is known as its proteome.

The chief characteristic of proteins that enables them to carry out their diverse cellular functions is their ability to bind other molecules specifically and tightly. The region of the protein responsible for binding another molecule is known as the binding site and is often a depression or "pocket" on the molecular surface. This binding ability is mediated by the tertiary structure of the protein, which defines the binding site pocket, and by the chemical properties of the surrounding amino acids' side chains. Protein binding can be extraordinarily tight and specific; for example, the ribonuclease inhibitor protein binds to human angiogenin with a sub-femtomolar dissociation constant (<10-15 M) but does not bind at all to its amphibian homolog onconase (>1 M). Extremely minor chemical changes such as the addition of a single methyl group to a binding partner can sometimes suffice to nearly eliminate binding; for example, the aminoacyl tRNA synthetase specific to the amino acid valine discriminates against the very similar side chain of the amino acid isoleucine.
Proteins can bind to other proteins as well as to small-molecule substrates. When proteins bind specifically to other copies of the same molecule, they can oligomerize to form fibrils; this process occurs often in structural proteins that consist of globular monomers that self-associate to form rigid fibers. Protein-protein interactions also regulate enzymatic activity, control progression through the cell cycle, and allow the assembly of large protein complexes that carry out many closely related reactions with a common biological function. Proteins can also bind to, or even be integrated into, cell membranes. The ability of binding partners to induce conformational changes in proteins allows the construction of enormously complex signaling networks.
Enzymes
The best-known role of proteins in the cell is their duty as enzymes, which catalyze chemical reactions. Enzymes are usually highly specific catalysts that accelerate only one or a few chemical reactions. Enzymes effect most of the reactions involved in metabolism and catabolism as well as DNA replication, DNA repair, and RNA synthesis. Some enzymes act on other proteins to add or remove chemical groups in a process known as post-translational modification. About 4,000 reactions are known to be catalyzed by enzymes.[14] The rate acceleration conferred by enzymatic catalysis is often enormous - as much as 1017-fold increase in rate over the uncatalyzed reaction in the case of orotate decarboxylase (78 million years without the enzyme, 18 milliseconds with the enzyme).[15]
The molecules bound and acted upon by enzymes are known as substrates. Although enzymes can consist of hundreds of amino acids, it is usually only a small fraction of the residues that come in contact with the substrate and an even smaller fraction - 3-4 residues on average - that are directly involved in catalysis.[16] The region of the enzyme that binds the substrate and contains the catalytic residues is known as the active site.
Cell signalling and ligand transport

Many proteins are involved in the process of cell signaling and signal transduction. Some proteins, such as insulin, are extracellular proteins that transmit a signal from the cell in which they were synthesized to other cells in distant tissues. Others are membrane proteins that act as receptors whose main function is to bind a signaling molecule and induce a biochemical response in the cell. Many receptors have a binding site exposed on the cell surface and an effector domain within the cell, which may have enzymatic activity or may undergo a conformational change detected by other proteins within the cell.
Antibodies are protein components of adaptive immune system whose main function is to bind antigens, or foreign substances in the body, and target them for destruction. Antibodies can be secreted into the extracellular environment or anchored in the membranes of specialized B cells known as plasma cells. While enzymes are limited in their binding affinity for their substrates by the necessity of conducting their reaction, antibodies have no such constraints. An antibody's binding affinity to its target is extraordinarily high.
Many ligand transport proteins bind particular small biomolecules and transport them to other locations in the body of a multicellular organism. These proteins must have a high binding affinity when their ligand is present in high concentrations but must also release the ligand when it is present at low concentrations in the target tissues. The canonical example of a ligand-binding protein is haemoglobin, which transports oxygen from the lungs to other organs and tissues in all vertebrates and has close homologs in every biological kingdom.
Transmembrane proteins can also serve as ligand transport proteins that alter the permeability of the cell's membrane to small molecules and ions. The membrane alone has a hydrophobic core through which polar or charged molecules cannot diffuse. Membrane proteins contain internal channels that allow such molecules to enter and exit the cell. Many ion channel proteins are specialized to select for only a particular ion; for example, potassium and sodium channels often discriminate for only one of the two ions.
Structural proteins
Structural proteins confer stiffness and rigidity to otherwise fluid biological components. Most structural proteins are fibrous proteins; for example, actin and tubulin are globular and soluble as monomers but polymerize to form long, stiff fibers that comprise the cytoskeleton, which allows the cell to maintain its shape and size. Collagen and elastin are critical components of connective tissue such as cartilage, and keratin is found in hard or filamentous structures such as hair, nails, feathers, hooves, and some animal shells.
Other proteins that serve structural functions are motor proteins such as myosin, kinesin, and dynein, which are capable of generating mechanical forces. These proteins are crucial for cellular motility of single-celled organisms and the sperm of many sexually reproducing multicellular organisms. They also generate the forces exerted by contracting muscles.
Methods of study
As some of the most commonly studied biological molecules, the activities and structures of proteins are examined both in vitro and in vivo. In vitro studies of purified proteins in controlled environments are useful for learning how a protein carries out its function: for example, enzyme kinetics studies explore the chemical mechanism of an enzyme's catalytic activity and its relative affinity for various possible substrate molecules. By contrast, in vivo experiments on proteins' activities within cells or even within whole organisms can provide complementary information about where a protein functions and how it is regulated.
Protein purification
In order to perform in vitro analyses, a protein must be purified away from other cellular components. This process usually begins with cell lysis, in which a cell's membrane is disrupted and its internal contents released into a solution known as a crude lysate. The resulting mixture can be purified using ultracentrifugation, which fractionates the various cellular components into fractions containing soluble proteins; membrane lipids and proteins; cellular organelles, and nucleic acids. Precipitation by a method known as salting out can concentrate the proteins from this lysate. Various types of chromatography are then used to isolate the protein or proteins of interest based on properties such as molecular weight, net charge and binding affinity. The level of purification can be monitored using gel electrophoresis if the desired protein's molecular weight is known, by spectroscopy if the protein has distinguishable spectroscopic features, or by enzyme assays if the protein has enzymatic activity.
For natural proteins, a series of purification steps may be necessary to obtain protein sufficiently pure for laboratory applications. To simplify this process, genetic engineering is often used to add chemical features to proteins that make them easier to purify without affecting their structure or activity. Here, a "tag" consisting of a specific amino acid sequence, often a series of histidine residues (a "His-tag"), is attached to one terminus of the protein. As a result, when the lysate is passed over a chromatography column containing nickel, the histidine residues ligate the nickel and attach to the column while the untagged components of the lysate pass unimpeded.
Cellular localization
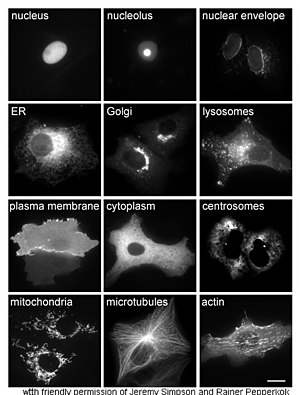
The study of proteins in vivo is often concerned with the synthesis and localization of the protein within the cell. Although many intracellular proteins are synthesized in the cytoplasm and membrane-bound or secreted proteins in the endoplasmic reticulum, the specifics of how proteins are targeted to specific organelles or cellular structures is often unclear. A useful technique for assessing cellular localization uses genetic engineering to express in a cell a fusion protein or chimera consisting of the natural protein of interest linked to a "reporter" such as green fluorescent protein (GFP). The fused protein's position within the cell can be cleanly and efficiently visualized using microscopy, as shown in the figure opposite.
Through another genetic engineering application known as site-directed mutagenesis, researchers can alter the protein sequence and hence its structure, cellular localization, and susceptibility to regulation, which can be followed in vivo by GFP tagging or in vitro by enzyme kinetics and binding studies.
Proteomics and bioinformatics
The total complement of proteins present at a time in a cell or cell type is known as its proteome, and the study of such large-scale data sets defines the field of proteomics, named by analogy to the related field of genomics. Key experimental techniques in proteomics include protein microarrays, which allow the detection of the relative levels of a large number of proteins present in a cell, and two-hybrid screening, which allows the systematic exploration of protein-protein interactions. The total complement of biologically possible such interactions is known as the interactome. A systematic attempt to determine the structures of proteins representing every possible fold is known as structural genomics.
The large amount of genomic and proteomic data available for a variety of organisms, including the human genome, allows researchers to efficiently identify homologous proteins in distantly related organisms by sequence alignment. Sequence profiling tools can perform more specific sequence manipulations such as restriction enzyme maps, open reading frame analyses for nucleotide sequences, and secondary structure prediction. From this data phylogenetic trees can be constructed and evolutionary hypotheses developed using special software like ClustalW regarding the ancestry of modern organisms and the genes they express. The field of bioinformatics seeks to assemble, annotate, and analyze genomic and proteomic data, applying computational techniques to biological problems such as gene finding and cladistics.
Structure prediction and simulation
Complementary to the field of structural genomics, protein structure prediction seeks to develop efficient ways to provide plausible models for proteins whose structures have not yet been determined experimentally. The most successful type of structure prediction, known as homology modeling, relies on the existence of a "template" structure with sequence similarity to the protein being modeled; structural genomics' goal is to provide sufficient representation in solved structures to model most of those that remain. Although producing accurate models remains a challenge when only distantly related template structures are available, it has been suggested that sequence alignment is the bottleneck in this process, as quite accurate models can be produced if a "perfect" sequence alignment is known.[17] Many structure prediction methods have served to inform the emerging field of protein engineering, in which novel protein folds have already been designed.[18] A more complex computational problem is the prediction of intermolecular interactions, such as in molecular docking and protein-protein interaction prediction.
The processes of protein folding and binding can be simulated using techniques derived from molecular dynamics, which increasingly take advantage of distributed computing as in the Folding@Home project. The folding of small alpha-helical protein domains such as the villin headpiece[19] and the HIV accessory protein[20] have been successfully simulated in silico, and hybrid methods that combine standard molecular dynamics with quantum mechanics calculations have allowed exploration of the electronic states of rhodopsins.[21]
Visualization
- PyMOL
- Sirius
- Rasmol
- Visual molecular dynamics
Nutrition
- Further information: Protein in nutrition
Most microorganisms and plants can biosynthesize all 20 standard amino acids, while animals must obtain some of the amino acids from the diet.[13] Key enzymes in the biosynthetic pathways that synthesize certain amino acids - such as aspartokinase, which catalyzes the first step in the synthesis of lysine, methionine, and threonine from aspartate - are not present in animals. The amino acids that an organism cannot synthesize on its own are referred to as essential amino acids. (This designation is often used to specifically identify those essential to humans.) If amino acids are present in the environment, most microorganisms can conserve energy by taking up the amino acids from the environment and downregulating their own biosynthetic pathways. Bacteria are often engineered in the laboratory to lack the genes necessary for synthesizing a particular amino acid, providing a selectable marker for the success of transfection, or the introduction of foreign DNA.
In animals, amino acids are obtained through the consumption of foods containing protein. Ingested proteins are broken down through digestion, which typically involves denaturation of the protein through exposure to acid and degradation by the action of enzymes called proteases. Ingestion of essential amino acids is critical to the health of the organism, since the biosynthesis of proteins that include these amino acids is inhibited by their low concentration. Amino acids are also an important dietary source of nitrogen. Some ingested amino acids, especially those that are not essential, are not used directly for protein biosynthesis. Instead, they are converted to carbohydrates through gluconeogenesis, which is also used under starvation conditions to generate glucose from the body's own proteins, particularly those found in muscle.
History
- Further information: History of molecular biology
Proteins were recognized as a distinct class of biological molecules in the eighteenth century by Antoine Fourcroy and others, distinguished by the molecules' ability to coagulate or flocculate under treatments with heat or acid. Noted examples at the time included albumen from egg whites, blood, serum albumin, fibrin, and wheat gluten. Dutch chemist Gerhardus Johannes Mulder carried out elemental analysis of common proteins and found that nearly all proteins had the same empirical formula. The term "protein" to describe these molecules was proposed in 1838 by Mulder's associate Jöns Jakob Berzelius. Mulder went on to identify the products of protein degradation such as the amino acid leucine for which he found a (nearly correct) molecular weight of 131 Da.
The difficulty in purifying proteins in large quantities made them very difficult for early protein biochemists to study. Hence, early studies focused on proteins that could be purified in large quantities, e.g., those of blood, egg white, various toxins, and digestive/metabolic enzymes obtained from slaughterhouses. In the late 1950s, the Armour Hot Dog Co. purified 1 kg (= one million milligrams) of pure bovine pancreatic ribonuclease A and made it freely available to scientists around the world.
Linus Pauling is credited with the successful prediction of regular protein secondary structures based on hydrogen bonding, an idea first put forth by William Astbury in 1933. Later work by Walter Kauzman on denaturation, based partly on previous studies by Kaj Linderstrom-Lang, contributed an understanding of protein folding and structure mediated by hydrophobic interactions. In 1949 Fred Sanger correctly determined the amino acid sequence of insulin, thus conclusively demonstrating that proteins consisted of linear polymers of amino acids rather than branched chains, colloids, or cyclols. The first atomic-resolution structures of proteins were solved by X-ray crystallography in the 1960s and by NMR in the 1980s. As of 2006, the Protein Data Bank has nearly 40,000 atomic-resolution structures of proteins. In more recent times, cryo-electron microscopy of large macromolecular assemblies and computational protein structure prediction of small protein domains are two methods approaching atomic resolution.
See also
- Amino acid
- Essential amino acid
- Protein design
- Intein
- List of recombinant proteins
- List of proteins
- Prion
- Edible protein per unit area of land
- Expression cloning
References
- ^ Sumner, JB (1926). "The Isolation and Crystallization of the Enzyme Urease. Preliminary Paper". J Biol Chem 69: 435-41.
- ^ Muirhead H, Perutz M (1963). "Structure of haemoglobin. A three-dimensional fourier synthesis of reduced human haemoglobin at 5.5 A resolution". Nature 199 (4894): 633-8. PMID 14074546.
- ^ Kendrew J, Bodo G, Dintzis H, Parrish R, Wyckoff H, Phillips D (1958). "A three-dimensional model of the myoglobin molecule obtained by x-ray analysis". Nature 181 (4610): 662-6. PMID 13517261.
- ^ Nelson, D. L. and Cox, M. M. (2005) Lehninger's Principles of Biochemistry, 4th Edition, W. H. Freeman and Company, New York.
- ^ a b c Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipurksy SL, Darnell J. (2004). Molecular Cell Biology 5th ed. WH Freeman and Company: New York, NY.
- ^ Dobson CM. (2000). The nature and significance of protein folding. In Mechanisms of Protein Folding 2nd ed. Ed. RH Pain. Frontiers in Molecular Biology series. Oxford University Press: New York, NY.
- ^ Fulton A, Isaacs W (1991). "Titin, a huge, elastic sarcomeric protein with a probable role in morphogenesis". Bioessays 13 (4): 157-61. PMID 1859393.
- ^ Bruckdorfer T, Marder O, Albericio F (2004). "From production of peptides in milligram amounts for research to multi-tons quantities for drugs of the future". Curr Pharm Biotechnol 5 (1): 29-43. PMID 14965208.
- ^ Schwarzer D, Cole P (2005). "Protein semisynthesis and expressed protein ligation: chasing a protein's tail". Curr Opin Chem Biol 9 (6): 561-9. PMID 16226484.
- ^ a b Branden C, Tooze J. (1999). Introduction to Protein Structure 2nd ed. Garland Publishing: New York, NY
- ^ Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. (2005). Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438(7068):633-8.
- ^ Walian P, Cross TA, Jap BK. (2004). Structural genomics of membrane proteins Genome Biol 5(4): 215.
- ^ a b Voet D, Voet JG. (2004). Biochemistry Vol 1 3rd ed. Wiley: Hoboken, NJ.
- ^ Bairoch A. (2000). "The ENZYME database in 2000". Nucleic Acids Res 28: 304-305. PMID 10592255.
- ^ Radzicka A, Wolfenden R. (1995). "A proficient enzyme.". Science 6 (267): 90-931. PMID 7809611.
- ^ The Catalytic Site Atlas at The European Bioinformatics Institute
- ^ Zhang Y, Skolnick J. (2005). The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci USA 102(4):1029-34.
- ^ Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. (2003). Design of a novel globular protein fold with atomic-level accuracy. Science 302(5649):1364-8.
- ^ Zagrovic B, Snow CD, Shirts MR, Pande VS. (2002). Simulation of folding of a small alpha-helical protein in atomistic detail using worldwide-distributed computing. J Mol Biol 323(5):927-37.
- ^ Herges T, Wenzel W. (2005). In silico folding of a three helix protein and characterization of its free-energy landscape in an all-atom force field. Phys Rev Let 94(1):018101.
- ^ Hoffmann M, Wanko M, Strodel P, Konig PH, Frauenheim T, Schulten K, Thiel W, Tajkhorshid E, Elstner M. (2006). Color tuning in rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J Am Chem Soc 128(33):10808-18.
External links
- Proteins (the journal), also called "Proteins: Structure, Function, and Bioinformatics" and previously "Proteins: Structure, Function, and Genetics" (1986-1995).
Databases and projects
- The Protein Databank (see also PDB Molecule of the Month, presenting short accounts on selected proteins from the PDB)
- UniProt the Universal Protein Resource
- Human Protein Atlas
- iHOP - Information Hyperlinked over Proteins
- MIT's Laboratory for Protein Molecular Self-Assembly
- NCBI Entrez Protein database
- NCBI Protein Structure database
- Human Protein Reference Database
- Human Proteinpedia
- Folding@Home (Stanford University)
Tutorials and educational websites
- Proteins: Biogenesis to Degradation - The Virtual Library of Biochemistry and Cell Biology
- Amino acid metabolism
- Data Book of Molecules - Home Page for Learning Environmental Chemistry